A human model of asthma exacerbation reveals transcriptional programs and cell circuits specific to allergic asthma
Jehan Alladina1,2*, Neal P Smith2,3,4*, Tristan Kooistra1,2, Kamil Slowikowski2,3,4, Isabela J Kernin2,3,4, Jacques Deguine3, Henry L Keen5, Kasidet Manakongtreecheep2,3,4, Jessica Tantivit2,3,4, Rod A Rahimi1,2, Susan L Sheng1, Nhan D Nguyen1,2, Alexis M Haring1,2, Francesca L Giacona1,2, Lida P Hariri1,6, Ramnik J Xavier3,7,8, Andrew D Luster2,3,9, Alexandra-Chloé Villani2,3,4‡, Josalyn L Cho10‡, Benjamin D Medoff1,2‡
Author affiliations
- 1 - Division of Pulmonary and Critical Care Medicine, Massachusetts General Hospital and Harvard Medical School, Boston, MA, USA.
- 2 - Center for Immunology and Inflammatory Diseases, Massachusetts General Hospital and Harvard Medical School, Boston, MA, USA.
- 3 - Broad Institute of Massachusetts Institute of Technology and Harvard, Cambridge, MA, USA.
- 4 - Massachusetts General Hospital Cancer Center, Boston, MA, USA.
- 5 - Iowa Institute of Human Genetics, University of Iowa Carver College of Medicine, Iowa City, IA, USA.
- 6 - Department of Pathology, Massachusetts General Hospital, Boston, MA, USA.
- 7 - Center for Computational and Integrative Biology, Massachusetts General Hospital and Harvard Medical School, Boston, MA, USA.
- 8 - Division of Gastroenterology, Massachusetts General Hospital and Harvard Medical School, Boston, MA, USA.
- 9 - Division of Rheumatology, Allergy and Immunology, Massachusetts General Hospital and Harvard Medical School, Boston, MA, USA.
- 10 - Division of Pulmonary, Critical Care and Occupational Medicine, University of Iowa Carver College of Medicine, Iowa City, IA, USA.
* These authors contributed equally: Molly Fisher Thomas, Kamil Slowikowski
‡These authors contributed equally: Alexnandra-Chloé Villani, Josalyn L Cho, Benjamin D Medoff
Science Immunology 2023. doi: 10.1126/sciimmunol.abq6352
Abstract
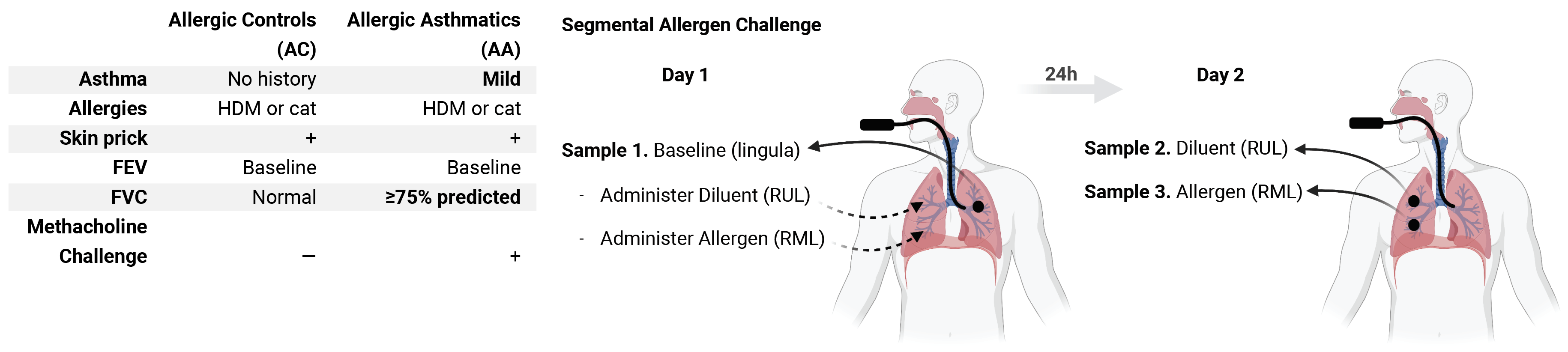
Using a human model of localized asthma exacerbation, we compared the lower airway mucosa in allergic asthmatics and allergic non-asthmatic controls using single-cell RNA-sequencing. In response to allergen challenge, the airway epithelium in asthmatics was highly dynamic and upregulated genes involved in matrix degradation, mucus metaplasia, and glycolysis while failing to induce injury-repair and antioxidant pathways observed in controls. IL9-expressing pathogenic TH2 cells were specific to asthmatic airways and were only observed after allergen challenge. Additionally, DC2 (CD1C) and CCR2-expressing monocyte-derived cells (MC) were uniquely enriched in asthmatics after allergen, with upregulation of genes that sustain type 2 inflammation and promote pathologic airway remodeling. In contrast, controls were enriched for macrophage-like MC that upregulated tissue repair programs after allergen challenge, suggesting these populations may protect against asthmatic airway remodeling. Cellular interaction analyses revealed a TH2-mononuclear phagocyte-epithelial interactome unique to asthmatics. These pathogenic airway circuits were characterized by type 2 programming of immune and structural cells, along with additional pathways that may sustain and amplify type 2 signals including TNF family signaling, altered cellular metabolism, failure to engage antioxidant responses, and the loss of growth factor signaling. Our findings therefore suggest that pathogenic effector circuits and the absence of pro-resolution programs drive structural airway disease in response to type 2 inflammation.
🔬 View the data
On this website, we provide interactive data browsers to view all of the transcriptomics data for each of the manually curated cell clusters.
Cell Clusters
Metadata variables and gene expression in two-dimensional embeddings.
3 cell lineages:
- Epithelial cells, Mononuclear phagocytes, T cells
Gene Contrasts
Differential expression statistics for all genes across:
- 4 contrasts (Ag vs Bln within AA or AC, AA vs AC within Ag or Bln)
📝 Cite our work
- Alladina J, Smith NP, Kooistra T, Slowikowski K, Kernin IJ, Deguine J, et al. A human model of asthma exacerbation reveals transcriptional programs and cell circuits specific to allergic asthma. Sci Immunol. 2023;8: eabq6352. doi:10.1126/sciimmunol.abq6352
BibTex
@ARTICLE{Alladina2023,
title = "{A human model of asthma exacerbation reveals transcriptional
programs and cell circuits specific to allergic asthma}",
author = "Alladina, Jehan and Smith, Neal P and Kooistra, Tristan and
Slowikowski, Kamil and Kernin, Isabela J and Deguine, Jacques and
Keen, Henry L and Manakongtreecheep, Kasidet and Tantivit,
Jessica and Rahimi, Rod A and Sheng, Susan L and Nguyen, Nhan D
and Haring, Alexis M and Giacona, Francesca L and Hariri, Lida P
and Xavier, Ramnik J and Luster, Andrew D and Villani,
Alexandra-Chlo{\'e} and Cho, Josalyn L and Medoff, Benjamin D",
abstract = "Asthma is a chronic disease most commonly associated with allergy
and type 2 inflammation. However, the mechanisms that link airway
inflammation to the structural changes that define asthma are
incompletely understood. Using a human model of allergen-induced
asthma exacerbation, we compared the lower airway mucosa in
allergic asthmatics and allergic non-asthmatic controls using
single-cell RNA sequencing. In response to allergen, the
asthmatic airway epithelium was highly dynamic and up-regulated
genes involved in matrix degradation, mucus metaplasia, and
glycolysis while failing to induce injury-repair and antioxidant
pathways observed in controls. IL9-expressing pathogenic TH2
cells were specific to asthmatic airways and were only observed
after allergen challenge. Additionally, conventional type 2
dendritic cells (DC2 that express CD1C) and CCR2-expressing
monocyte-derived cells (MCs) were uniquely enriched in asthmatics
after allergen, with up-regulation of genes that sustain type 2
inflammation and promote pathologic airway remodeling. In
contrast, allergic controls were enriched for macrophage-like MCs
that up-regulated tissue repair programs after allergen
challenge, suggesting that these populations may protect against
asthmatic airway remodeling. Cellular interaction analyses
revealed a TH2-mononuclear phagocyte-basal cell interactome
unique to asthmatics. These pathogenic cellular circuits were
characterized by type 2 programming of immune and structural
cells and additional pathways that may sustain and amplify type 2
signals, including TNF family signaling, altered cellular
metabolism, failure to engage antioxidant responses, and loss of
growth factor signaling. Our findings therefore suggest that
pathogenic effector circuits and the absence of proresolution
programs drive structural airway disease in response to type 2
inflammation.",
journal = "Science immunology",
volume = 8,
number = 83,
pages = "eabq6352",
month = may,
year = 2023,
language = "en",
issn = "2470-9468",
pmid = "37146132",
doi = "10.1126/sciimmunol.abq6352",
pmc = "PMC10440046"
}
💾 Download the data
The single-cell data is available at NCBI GEO accession GSE193816.
💻 Read the source code
Analysis output files and source code for the analysis is available at GitHub: github.com/villani-lab/airway_allergic_asthma
✉ Contact us
Please contact us with any questions or comments.
The data presented here comes from the laboratories of Dr. Benjamin Medoff and Dr. Josalyn Cho.